Deep learning classification of apple leaf diseases: comparison of neural networks
Keywords:
Deep learning, Convolutional neural networks, Apple trees, Rust, Scab, EnsemblesAbstract
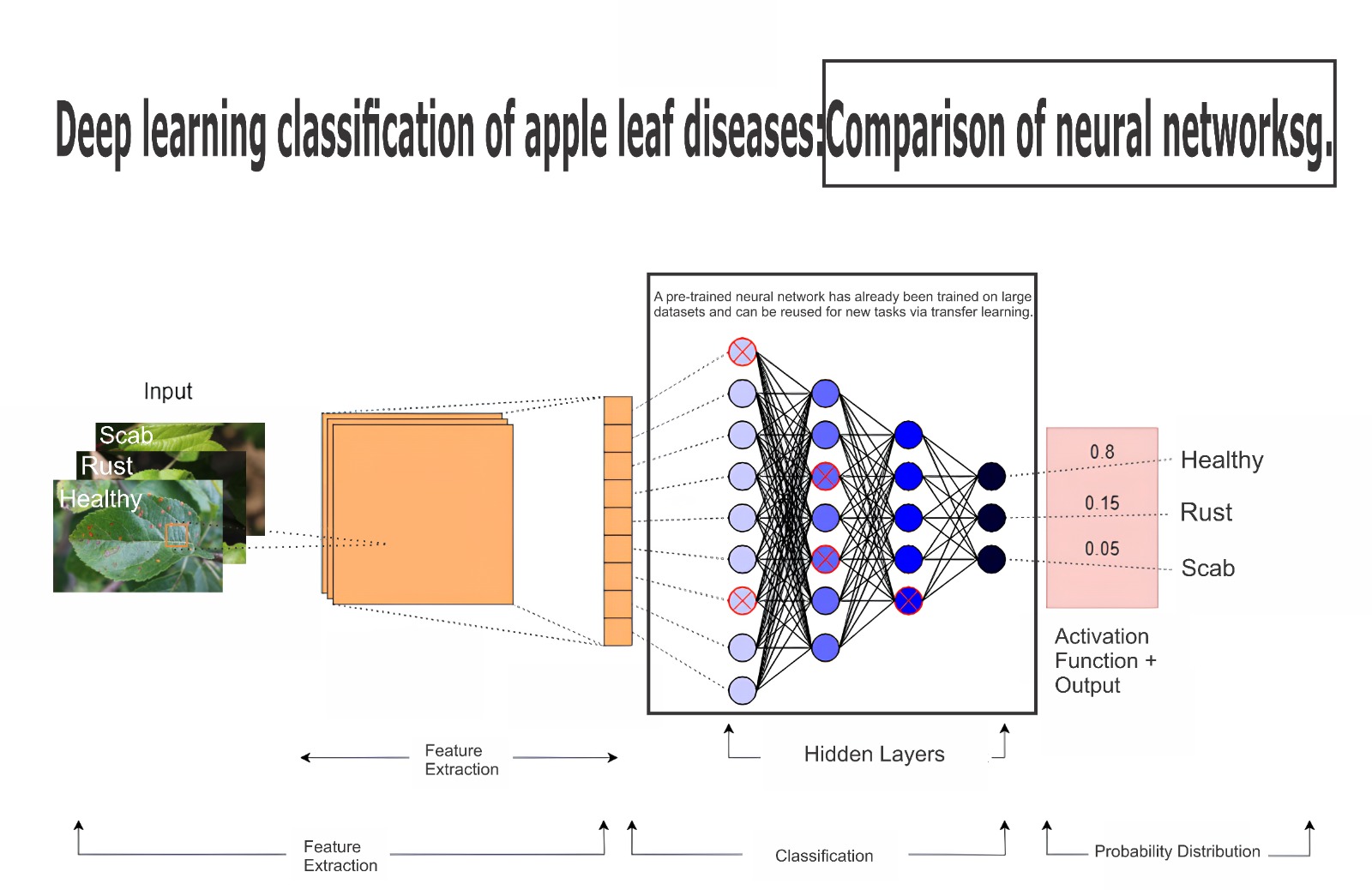
The production of apples is a significant segment of the global agricultural industry, often threatened by diseases and pests. This study investigates the use of convolutional neural networks (CNNs) to classify images of apple tree leaves, distinguishing between healthy leaves and those affected by rust and scab. The objective is to develop an approach for the early detection of these fungal diseases. High-resolution images were collected, considering variations in lighting, angles, and backgrounds. Eighteen pre-trained CNN architectures available in Keras were tested and evaluated using metrics such as accuracy, precision, recall, and F1-score. The EfficientNetV2B2 and DenseNet201 networks showed the best results, with an accuracy of 99%. To enhance classification performance, ensemble techniques were explored, including combining all networks and selecting only the most accurate ones. Although promising, challenges such as computational complexity and the need for real-time processing in practical applications remain. The findings demonstrate the potential of CNNs and ensemble methods in supporting early detection of diseases in apple orchards, providing valuable tools for producers to manage infestations more effectivelys.
References
AGROLINK. Sarna da maçã (venturia inaequalis). Portal Agrolink, 2020. Disponível em: <https://www.agrolink.com.br/problemas/sarna-da-maca_1692.html>.
ALSUWAT, M. et al. Prediction of diabetic retinopathy using convolutional neural networks. International Journal of Advanced Computer Science and Applications, v. 13, n. 7, 2022. Disponível em: <https://doi.org/10.14569/ijacsa.2022.0130798>.
ATLASBIG. Principais países produtores de maçã. 2021. Accessed on 2023-07-30. Disponível em: <https://www.atlasbig.com/pt-br/paises-por-producao-de-maca>.
CHENG, P.; MALHI, H. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. Journal of Digital Imaging, v. 30, n. 2, p. 234–243, 2016. Disponível em: <https://doi.org/10.1007/s10278-016-9929-2>.
CRUZ, A. D. Deep neural network approach for sentiment analysis of tweets related to sports concussion. 2023.
CULTIVAR. Sarna na maçã. Revista Cultivar, 2018. Disponível em: <https://revistacultivar.com.br/artigos/sarna-na-maca>.
CULTIVAR. Controle da sarna da macieira. Revista Cultivar, 2019. Disponível em:<https://revistacultivar.com.br/artigos/controle-da-sarna-da-macieira>.
FAO. World apple production in 2021/2022 reaches 86.5 million tonnes. 2021. Accessed on 2023-07-30. Disponível em: <https://www.fao.org/news/story/en/item/1425859/icode/>.
FU, L. et al. Lightweight-convolutional neural network for apple leaf disease identification. Frontiers in Plant Science, v. 13, 2022. Disponível em: <https://doi.org/10.3389/fpls.2022.831219>.
GIRSHICK, R. et al. Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition. [s.n.], 2014. Disponível em: <https://doi.org/10.1109/cvpr.2014.81>.
GUPTA, N.; SLAWSON, D. D.; MOFFAT, A. J. Using citizen science for early detection of tree pests and diseases: perceptions of professional and public participants. Biological Invasions, v. 24, n. 1, p. 123–138, 2022.
HE, K. et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
HU, F. et al. Transferring deep convolutional neural networks for the scene classification of high-resolution remote sensing imagery. Remote Sensing, v. 7, n. 11, p. 14680–14707, 2015. Disponível em: <https://doi.org/10.3390/rs71114680>.
IMAGENET. ImageNet Large Scale Visual Recognition Challenge (LSVRC). <https://www.image-net.org/challenges/LSVRC/>. Acesso em 1o de agosto de 2023.
JUNIOR, A. M. M.; SANTOS, P. R.; SAFADI, T. Utilização de redes neurais artificiais na classificação de danos em sementes de girassol. Sigmae, v. 8, n. 2, p. 564–575, 2019. Accessed: June 24, 2024. Disponível em: <https://publicacoes.unifal-mg.edu.br/revistas/index.php/sigmae/article/view/1046>.
KAGGLE. Plant Pathology 2020 - FGVC7. 2020. <https://www.kaggle.com/competitions/plant-pathology-2020-fgvc7/overview>. Accessed: 2023-07-31.
KOVALENKO, I. Redes neurais profundas (parte vi). ensemble de classificadores de redes neurais. MQL5, 2018. Disponível em: <https://www.mql5.com/pt/articles/4227>.
LEE, H.; HONG, S.; KIM, E. A new genetic feature selection with neural network ensemble. International Journal of Computer Mathematics, v. 86, p. 1105–1117, 2009.
LUDOVICO, S. N. et al. Agricultural commodity price prediction via machine learning algorithms. Sigmae, v. 11, n. 2, p. 45–69, 2023. Acesso em: 24 jun. 2024. Disponível em:<https://publicacoes.unifal-mg.edu.br/revistas/index.php/sigmae/article/view/1967>.
MA, H. et al. Integrating growth and environmental parameters to discriminate powdery mildew and aphid of winter wheat using bi-temporal landsat-8 imagery. Remote Sensing, v. 11, n. 7, p. 846, 2019. Disponível em: <https://doi.org/10.3390/rs11070846>.
MOHANTY, S.; HUGHES, D.; SALATHE, M. Using deep learning for image-based plant disease detection. Frontiers in Plant Science, v. 7, 2016. Disponível em: <https://doi.org/10.3389/fpls.2016.01419>.
OPITZ, D. W.; MACLIN, R. Popular ensemble methods: an empirical study. Journal of Artificial Intelligence Research, v. 11, p. 169–198, 1999.
PEIL, . W. A. The role of citizen science in the detection of invasive species. Biological Invasions, v. 11, n. 12, p. 2693–2707, 2009.
SCIARRETTA, A.; TREMATERRA, P. Geostatistical tools for the study of insect spatial distribution: practical implications in the integrated management of orchard and vineyard pests. Plant Protection Science, v. 50, n. 2, p. 97–110, 2014. Disponível em <https://doi.org/10.17221/40/2013-pps>.
SILVA, J. et al. Visão computacional para detecção de doenças fúngicas na agricultura. Revista UNICA, v. 1, n. 1, p. 1–10, 2020. Disponível em: <http://co.unicaen.com.br:89/periodicos/index.php/UNICA/article/view/67>.
SILVA, J. C. et al. Avaliação de cultivares de milho para produção de silagem. Revista Brasileira de Milho e Sorgo, v. 9, n. 1, p. 6–7, 2010. Disponível em: <https://ainfo.cnptia.embrapa.br/digital/bitstream/item/200689/1/12688-2010-p.6-7.pdf>.
SILVA, J. V. d. Estudo e análise de Redes Neurais Convolucionais para identificação de doenças em folhas de macieira. Dissertação (Mestrado) — Universidade de Brasília, 2021. Disponível em: <https://repositorio.unb.br/handle/10482/43328>.
SZEGEDY, C. et al. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015.
TAJBAKHSH, N. et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Transactions on Medical Imaging, v. 35, n. 5, p. 1299–1312, 2016. Disponível em: <https://doi.org/10.1109/tmi.2016.2535302>.
TECHINFUS. Por que aparece ferrugem na macieira e o que fazer? techinfus.com, 2021. Disponível em: <https://ibuilder-pt.techinfus.com/yabloni/rzhavchina/>.
TENSORFLOW. Instalação do TensorFlow com suporte a GPU (Português). <https://www.tensorflow.org/install/gpu?hl=pt-br>. Acesso em 1o de agosto de 2023.
THAPA, S. N. B. S. K. A. The plant pathology challenge 2020 data set to classify foliar disease of apples. Applications in Plant Sciences, v. 8, 2020.
TURKOGLU, M.; HANBAY, D.; SENGUR, A. Multi-model lstm-based convolutional neural networks for detection of apple diseases and pests. Journal of Ambient Intelligence and Humanized Computing, v. 13, n. 7, p. 3335–3345, 2019. Disponível em: <https://doi.org/10.1007/s12652-019-01591-w>.
VISHNOI, V. K.; KUMAR, K.; KUMAR, B. Plant disease detection using computational intelligence and image processing. Journal of Plant Disease Protection, v. 128, p. 19–53, 2021.
WANG, J. et al. Transferring pre-trained deep cnns for remote scene classification with general features learned from linear pca network. Remote Sensing, v. 9, n. 3, p. 225, 2017. Disponível em: <https://doi.org/10.3390/rs9030225>.
WANG, L. et al. Applications and prospects of agricultural unmanned aerial vehicle obstacle avoidance technology in china. Sensors, v. 19, n. 3, p. 642, 2019. Disponível em: <https://doi.org/10.3390/s19030642>.
WANG, P. et al. Identification of apple leaf diseases by improved deep convolutional neural networks with an attention mechanism. Frontiers in Plant Science, v. 12, 2021. Disponível em: <https://doi.org/10.3389/fpls.2021.723294>.
ZHANG, S. et al. Editorial: machine learning and artificial intelligence for smart agriculture, volume ii. Frontiers in Plant Science, v. 14, 2023. Disponível em: <https://doi.org/10.3389/fpls.2023.1166209>.
Downloads
Published
How to Cite
Issue
Section
License
Proposta de Política para Periódicos de Acesso Livre
Autores que publicam nesta revista concordam com os seguintes termos:
- Autores mantém os direitos autorais e concedem à revista o direito de primeira publicação, com o trabalho simultaneamente licenciado sob a Licença Creative Commons Attribution que permite o compartilhamento do trabalho com reconhecimento da autoria e publicação inicial nesta revista.
- Autores têm autorização para assumir contratos adicionais separadamente, para distribuição não-exclusiva da versão do trabalho publicada nesta revista (ex.: publicar em repositório institucional ou como capítulo de livro), com reconhecimento de autoria e publicação inicial nesta revista.
- Autores têm permissão e são estimulados a publicar e distribuir seu trabalho online (ex.: em repositórios institucionais ou na sua página pessoal) a qualquer ponto antes ou durante o processo editorial, já que isso pode gerar alterações produtivas, bem como aumentar o impacto e a citação do trabalho publicado (Veja O Efeito do Acesso Livre).



